Abstract
Introduction
Minimal residual disease (MRD) negativity after initial therapy is a strong predictor of survival in multiple myeloma. Tracking of clonal immunoglobulin V(D)J rearrangements by next generation sequencing is highly sensitive for MRD and does not require immediate analysis of fresh samples. However, previous studies have found variable rates of baseline V(D)J sequence capture, which could limit tracking. In this study, we aimed to define the sample-related and disease-related factors that influence V(D)J capture.
Methods
We included 177 patients with plasma cell myeloma who had available stored mononuclear cells from a baseline bone marrow aspirate. Each sample was sequenced by two assays: The LymphoTrack® VDJ assays from Invivoscribe, and our in-house myeloma panel myTYPE, as a molecular control for detectable tumor derived DNA in the samples. MyTYPE positivity was defined by one or more single nucleotide variant, insertion, deletion, translocation or copy number variation that is known to occur in myeloma.
Results and discussion
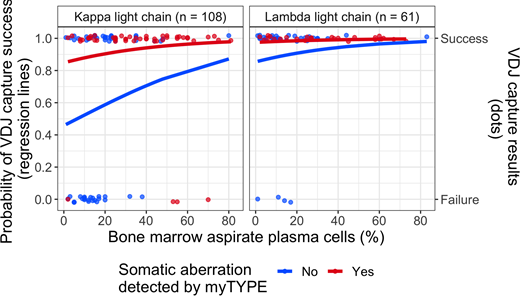
The V(D)J capture rate in our whole cohort was 81 %, as compared with 95 % in the myTYPE positive samples, demonstrating the importance of tumor cell content for V(D)J capture. This was confirmed in multivariate logistic regression (Figure 1), where myTYPE positivity was a strong independent predictor of V(D)J capture success, with an odds ratio (OR) of 6.61 (95 % CI 2.22-24.81, p = 0.002). Plasma cell content estimated from bone marrow aspirate smears also contributed to the multivariate model, with an OR of 1.3 for each 10 % increase in plasma cell content (95 % CI 0.96-1.84, p=0.109), but this did not reach statistical significance after accounting for the strong effect of myTYPE. Finally, having lambda light chain restricted plasma cells was a strong predictor of V(D)J capture success (OR 6.91, 95 % CI 2.4-25.32, p = 0.001).
Higher V(D)J capture rate in lambda-restricted myeloma as compared with kappa-restricted was mostly driven by a difference in immunoglobulin kappa gene (IGK) rearrangement capture (73 vs. 44 %, p < 0.001). As a potential explanation, we found up to 4 unique IGK rearrangements that are amenable to capture in lambda-restricted cases, as well as dramatically lower somatic hypermutation (SHM) of the IGK variable region in clonal rearrangements, as compared with kappa-restricted cases. SHM has previously been shown to cause V(D)J capture failure by interfering with PCR primer annealing. Both of these factors can be attributed to IGK inactivation by rearrangements involving the "kappa deleting element" region, affecting both IGK alleles in lambda-restricted plasma cells (Perfetti et al, Immunology, 2004).
As an explanation for low plasma cell content in the samples used in this study, we describe how the tumor cell content of bone marrow aspirates decrease gradually in sequential pulls because of hemodilution: from the initial pull used for aspirate smear, to the final pull that is commonly used for research. Supporting the important role of hemodilution, we found V(D)J capture rates of 97 % in clinical samples (early pull aspirates) from our institution that were analyzed with the same NGS assays, as long as the bone marrow plasma cell infiltration was above 5 %.
V(D)J capture probability appears to be determined by two factors: The abundance of clonal cells (i.e. tumor cell content), and the degree to which clonal sequences can be amplified by the assay (which is negatively affected by SHM). Thus, increasing the tumor cell content in samples as much as practically possible (i.e., optimal bone marrow aspirates and enrichment of CD138+ plasma cells) may compensate for SHM and improve V(D)J capture rates beyond 95 %.
Conclusion
V(D)J capture rates of at least 95 % are feasible in multiple myeloma using LymphoTrack® NGS assays, when the sample quality is good. The most important reason for V(D)J capture failure is low tumor cell content due to bone marrow aspirate hemodilution. Optimal performance depends on the use of early pull aspirates and/or subsequent tumor cell enrichment.
Figure 1: Predicting V(D)J capture. Regression lines and individual data points are colored according to myTYPE status (red = positive; blue = negative); V(D)J capture probability on the y-axis (capture yes/no on the second y-axis) and bone marrow plasma cell percentage by aspirate smear on the x-axis; split into panels according to light chain restriction (right = lambda; left = kappa).
Ho:Invivoscribe, Inc.: Honoraria. Arcila:Invivoscribe, Inc.: Consultancy, Honoraria. Jacobsen:Invivoscribe, Inc.: Employment. Huang:Invivoscribe, Inc.: Employment. Miller:Invivoscribe, Inc.: Employment, Equity Ownership. Landgren:Pfizer: Consultancy; Karyopharm: Consultancy; Amgen: Consultancy, Research Funding; Janssen: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Celgene: Consultancy, Research Funding; Merck: Membership on an entity's Board of Directors or advisory committees; Takeda: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal